Showing
- docs/source/tutorial.rst 0 additions, 59 deletionsdocs/source/tutorial.rst
- docs/source/workflow.rst 0 additions, 284 deletionsdocs/source/workflow.rst
- docs/source/xfel_calibrate_conf.rst 0 additions, 6 deletionsdocs/source/xfel_calibrate_conf.rst
- docs/static/calcat/CC_multiple_CCVs_calcat.png 0 additions, 0 deletionsdocs/static/calcat/CC_multiple_CCVs_calcat.png
- docs/static/calcat/home_calcat.png 0 additions, 0 deletionsdocs/static/calcat/home_calcat.png
- docs/static/calcat/home_cc_calcat.png 0 additions, 0 deletionsdocs/static/calcat/home_cc_calcat.png
- docs/static/calcat/home_ccv_calcat.png 0 additions, 0 deletionsdocs/static/calcat/home_ccv_calcat.png
- docs/static/calcat/home_conditions_calcat.png 0 additions, 0 deletionsdocs/static/calcat/home_conditions_calcat.png
- docs/static/calcat/home_detectors_calcat.png 0 additions, 0 deletionsdocs/static/calcat/home_detectors_calcat.png
- docs/static/calcat/home_pdu_calcat.png 0 additions, 0 deletionsdocs/static/calcat/home_pdu_calcat.png
- docs/static/calcat/home_reports_calcat.png 0 additions, 0 deletionsdocs/static/calcat/home_reports_calcat.png
- docs/static/detector_mapping/AGIPD_modules_pic.png 0 additions, 0 deletionsdocs/static/detector_mapping/AGIPD_modules_pic.png
- docs/static/detector_mapping/JUNGFRAU_PDU_pic.png 0 additions, 0 deletionsdocs/static/detector_mapping/JUNGFRAU_PDU_pic.png
- docs/static/detector_mapping/icalibrationdb_agipd_det_mapping.png 0 additions, 0 deletions...tic/detector_mapping/icalibrationdb_agipd_det_mapping.png
- docs/static/how_to_write_xfel_calibrate_notebook_NBC_14_0.png 0 additions, 0 deletions.../static/how_to_write_xfel_calibrate_notebook_NBC_14_0.png
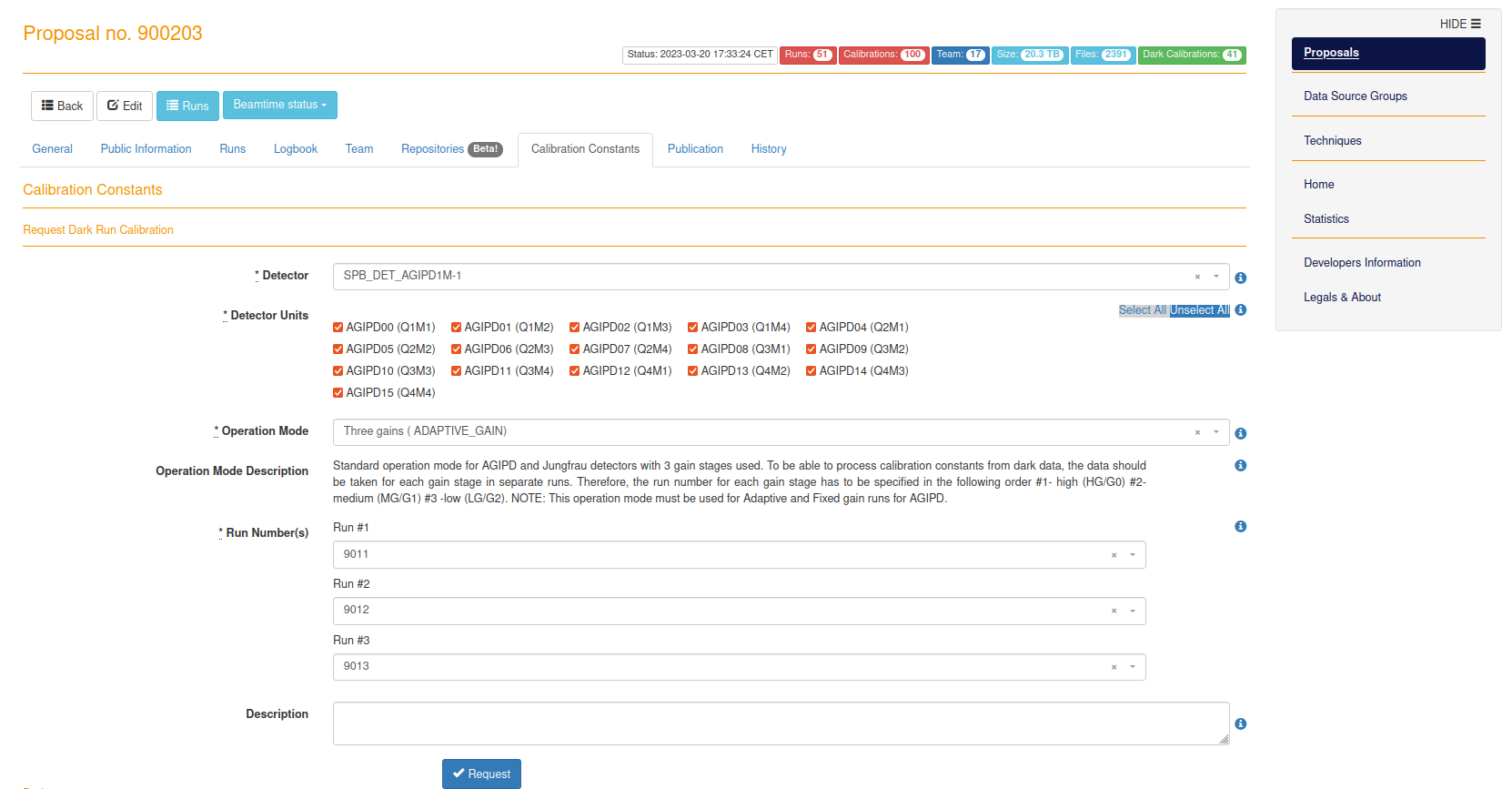
- docs/static/myMDC/calibration_constants.png 0 additions, 0 deletionsdocs/static/myMDC/calibration_constants.png
- docs/static/myMDC/correction.png 0 additions, 0 deletionsdocs/static/myMDC/correction.png
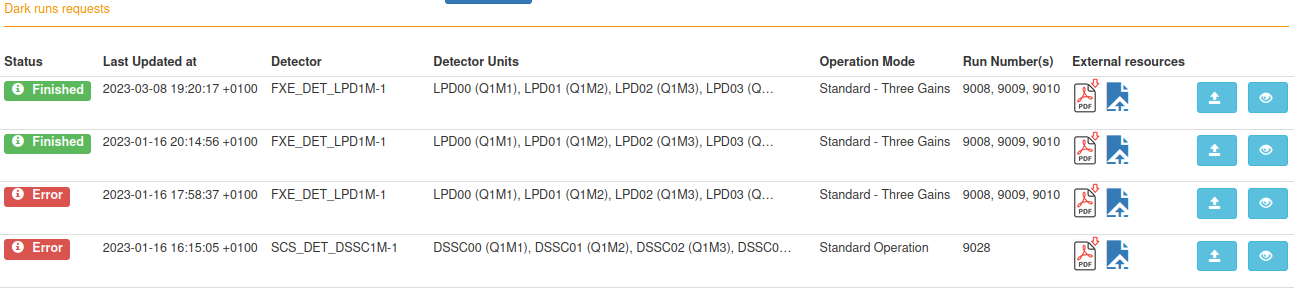
- docs/static/myMDC/dark_different_requests.png 0 additions, 0 deletionsdocs/static/myMDC/dark_different_requests.png
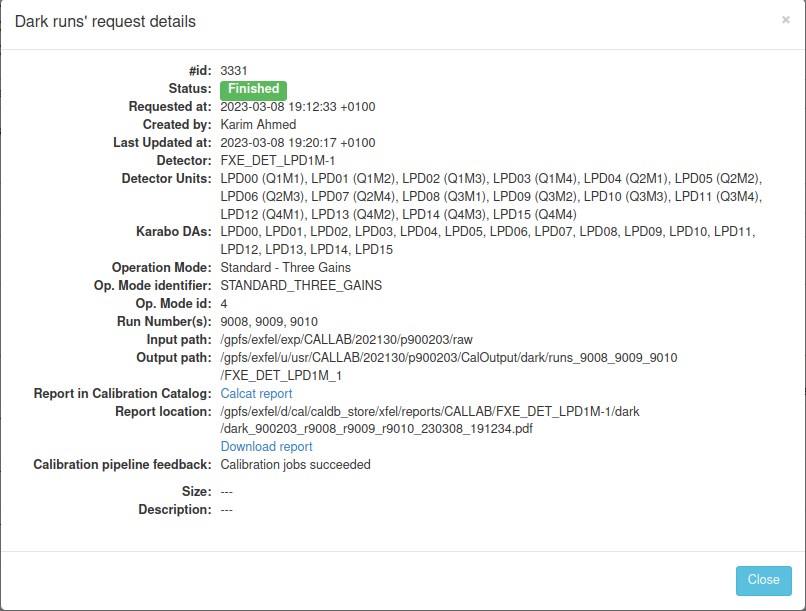
- docs/static/myMDC/dark_request_status_error.png 0 additions, 0 deletionsdocs/static/myMDC/dark_request_status_error.png
- docs/static/myMDC/dark_request_status_success.png 0 additions, 0 deletionsdocs/static/myMDC/dark_request_status_success.png
docs/source/tutorial.rst
deleted
100644 → 0
docs/source/workflow.rst
deleted
100644 → 0
docs/source/xfel_calibrate_conf.rst
deleted
100644 → 0
165 KiB
docs/static/calcat/home_calcat.png
0 → 100644
64.1 KiB
docs/static/calcat/home_cc_calcat.png
0 → 100644
134 KiB
docs/static/calcat/home_ccv_calcat.png
0 → 100644
182 KiB
85.6 KiB
docs/static/calcat/home_detectors_calcat.png
0 → 100644
92.7 KiB
docs/static/calcat/home_pdu_calcat.png
0 → 100644
122 KiB
docs/static/calcat/home_reports_calcat.png
0 → 100644
69.6 KiB
810 KiB
409 KiB
111 KiB
378 KiB
docs/static/myMDC/calibration_constants.png
0 → 100644
139 KiB
docs/static/myMDC/correction.png
0 → 100644
144 KiB
75.8 KiB
113 KiB
113 KiB