Showing
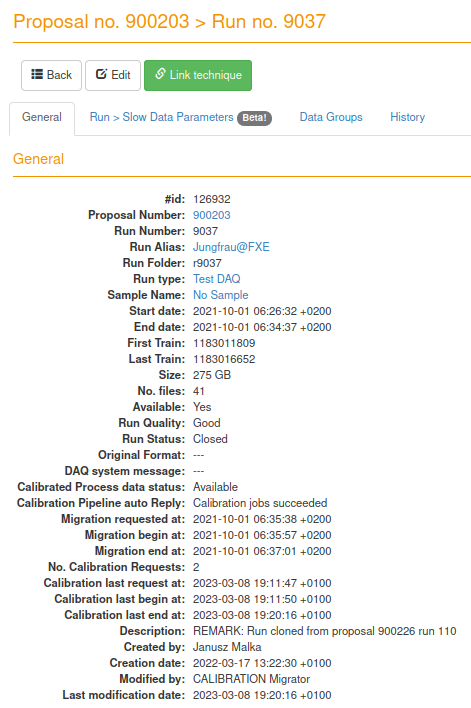
- docs/static/myMDC/run_9037_general_status.png 0 additions, 0 deletionsdocs/static/myMDC/run_9037_general_status.png
- docs/static/tests/given_argument_example.png 0 additions, 0 deletionsdocs/static/tests/given_argument_example.png
- docs/static/tests/manual_action.png 0 additions, 0 deletionsdocs/static/tests/manual_action.png
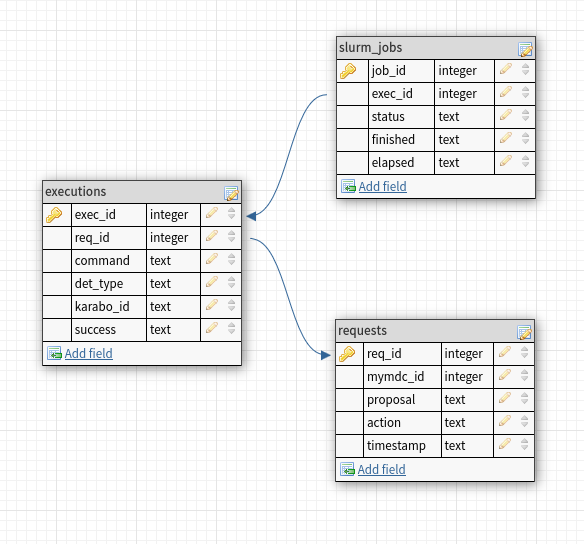
- docs/static/webservice_job_db.png 0 additions, 0 deletionsdocs/static/webservice_job_db.png
- docs/static/xfel_calibrate_diagrams/overview_all_services.png 0 additions, 0 deletions.../static/xfel_calibrate_diagrams/overview_all_services.png
- docs/static/xfel_calibrate_diagrams/xfel-calibrate_cli_process.png 0 additions, 0 deletions...ic/xfel_calibrate_diagrams/xfel-calibrate_cli_process.png
- mkdocs.yml 107 additions, 0 deletionsmkdocs.yml
- notebooks/AGIPD/AGIPD_Correct_and_Verify.ipynb 117 additions, 86 deletionsnotebooks/AGIPD/AGIPD_Correct_and_Verify.ipynb
- notebooks/AGIPD/Characterize_AGIPD_Gain_Darks_NBC.ipynb 30 additions, 29 deletionsnotebooks/AGIPD/Characterize_AGIPD_Gain_Darks_NBC.ipynb
- notebooks/AGIPD/Chracterize_AGIPD_Gain_PC_NBC.ipynb 48 additions, 160 deletionsnotebooks/AGIPD/Chracterize_AGIPD_Gain_PC_NBC.ipynb
- notebooks/AGIPD/Chracterize_AGIPD_Gain_PC_Summary.ipynb 726 additions, 0 deletionsnotebooks/AGIPD/Chracterize_AGIPD_Gain_PC_Summary.ipynb
- notebooks/DSSC/Characterize_DSSC_Darks_NBC.ipynb 1 addition, 3 deletionsnotebooks/DSSC/Characterize_DSSC_Darks_NBC.ipynb
- notebooks/Gotthard2/Characterize_Darks_Gotthard2_NBC.ipynb 138 additions, 79 deletionsnotebooks/Gotthard2/Characterize_Darks_Gotthard2_NBC.ipynb
- notebooks/Gotthard2/Correction_Gotthard2_NBC.ipynb 326 additions, 203 deletionsnotebooks/Gotthard2/Correction_Gotthard2_NBC.ipynb
- notebooks/Gotthard2/Summary_Darks_Gotthard2_NBC.ipynb 204 additions, 0 deletionsnotebooks/Gotthard2/Summary_Darks_Gotthard2_NBC.ipynb
- notebooks/Jungfrau/Jungfrau_Gain_Correct_and_Verify_NBC.ipynb 29 additions, 19 deletions...books/Jungfrau/Jungfrau_Gain_Correct_and_Verify_NBC.ipynb
- notebooks/Jungfrau/Jungfrau_dark_analysis_all_gains_burst_mode_NBC.ipynb 6 additions, 1 deletion...rau/Jungfrau_dark_analysis_all_gains_burst_mode_NBC.ipynb
- notebooks/Jungfrau/Jungfrau_darks_Summary_NBC.ipynb 70 additions, 35 deletionsnotebooks/Jungfrau/Jungfrau_darks_Summary_NBC.ipynb
- notebooks/LPD/LPDChar_Darks_NBC.ipynb 17 additions, 9 deletionsnotebooks/LPD/LPDChar_Darks_NBC.ipynb
- notebooks/LPDMini/LPD_Mini_Char_Darks_NBC.ipynb 26 additions, 15 deletionsnotebooks/LPDMini/LPD_Mini_Char_Darks_NBC.ipynb
86.3 KiB
docs/static/tests/given_argument_example.png
0 → 100644
40.3 KiB
docs/static/tests/manual_action.png
0 → 100644
87.1 KiB
docs/static/webservice_job_db.png
0 → 100644
59 KiB
48.8 KiB
34.5 KiB
mkdocs.yml
0 → 100644
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.
This diff is collapsed.